蒸汽量预测
案例来源:天池大赛——工业蒸汽量预测传送门 。
1 赛题背景
火力发电的基本原理 是:燃料在燃烧时加热水生成蒸汽,蒸汽压力推动汽轮机旋转,然后汽轮机带动发电机旋转,产生电能。在这一系列的能量转化中,影响发电效率的核心是锅炉的燃烧效率,即燃料燃烧加热水产生高温高压蒸汽。
锅炉的燃烧效率的影响因素 很多,包括锅炉的可调参数,如燃烧给量,一二次风,引风,返料风,给水水量;以及锅炉的工况,比如锅炉床温、床压,炉膛温度、压力,过热器的温度等。
2 赛题描述
经脱敏后的锅炉传感器采集数据(采集频率是分钟级别),根据锅炉的工况预测产生的蒸汽量。
3 数据说明
数据分成训练数据(train.txt)和测试数据(test.txt),其中字段”V0”-“V37”,这38个字段是作为特征变量,”target”作为目标变量。我们需要利用训练数据训练出模型,预测测试数据的目标变量,排名结果依据预测结果的MSE (mean square error)—— 评价指标。
4 案例分析和解决方案
4.1 导入包
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 import warningswarings.filterwarnings('ignore' ) import matplotlib.pyplot as pltimport seaborn as snsimport pandas as pdimport numpy as npfrom scipy import statsfrom sklearn.model_selection import train_test_splitfrom sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score, cross_val_predict, KFoldfrom sklearn.metrics import make_scorner, mean_squared_errorfrom sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNetfrom sklearn.svm import Linear_SVR, SVRfrom sklearn.neighbors import KNeighborsRegressionfrom sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor, AdaBoostRegressorfrom xgboost import XGBRegressorfrom sklearn.preprocessing import PolynomialFeatures, MinMaxScaler, StandardScaler
4.2 加载数据
1 2 data_train = pd.readcsv('train.txt' , sep='\t' ) data_test = pd.readcsv('test.txt' , sep='\t' )
1 2 3 4 5 6 data_train['oringin' ] = 'train' data_test['oringin' ] = 'test' data_all = pd.concat([data_train, data_test], axis=0 , ignore_index=True ) data_all.head()
4.3 探索数据分布
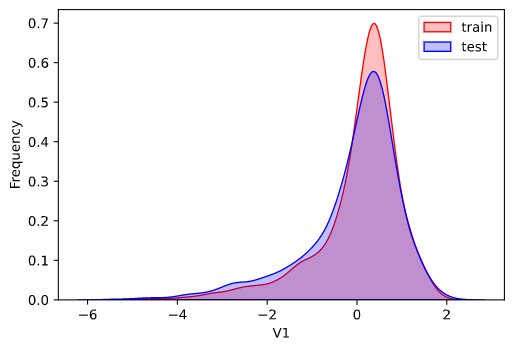
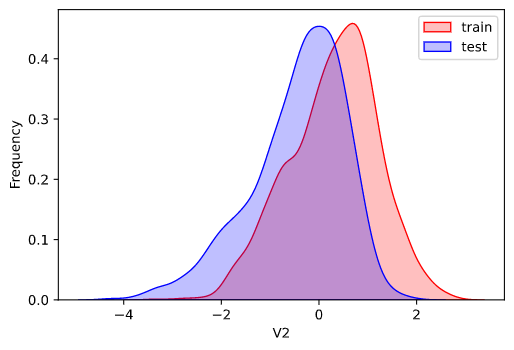
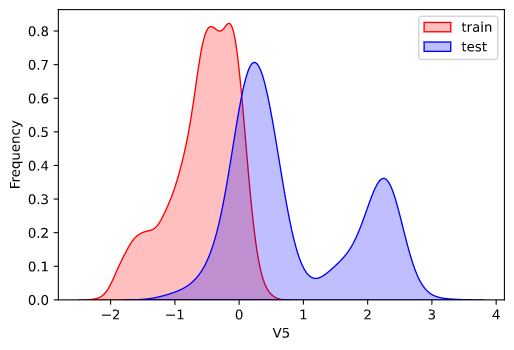
因为数据为传感器数据,即连续变量,所以使用核密度估计图(kdeplot)进行数据的初步分析(EDA:Exploratory data analysis)。
seaborn中的kdeplot函数参数、作用以及使用方法可以参考官方教程:传送门
1 2 3 4 5 6 7 8 for column in data_all.column[0 : -2 ]: g = sns.kdeplot(data_all[column][(data_all['oringin' ] == 'train' )], color='Red' , shade=True ) g = sns.kdeplot(data_all[column][(data_all['oringin' ] == 'test' )], ax=g, color='Blue' , shade=True ) g.set_xlabel(column) g.set_ylabel('Frequency' ) g = g.legend(['train' , 'test' ]) plt.show()
部分输出:
从上面的各特征概率分布函数曲线可以看出特征"V5","V9","V11","V17","V22"中训练集数据分布和测试集数据分布不均,所以我们删除这些特征数据。
1 2 3 4 5 6 7 8 9 10 data_all.drop(['V5' , 'V9' , 'V11' , 'V17' , 'V22' ], axis=1 , inplace=True ) data_all.head()
输出:
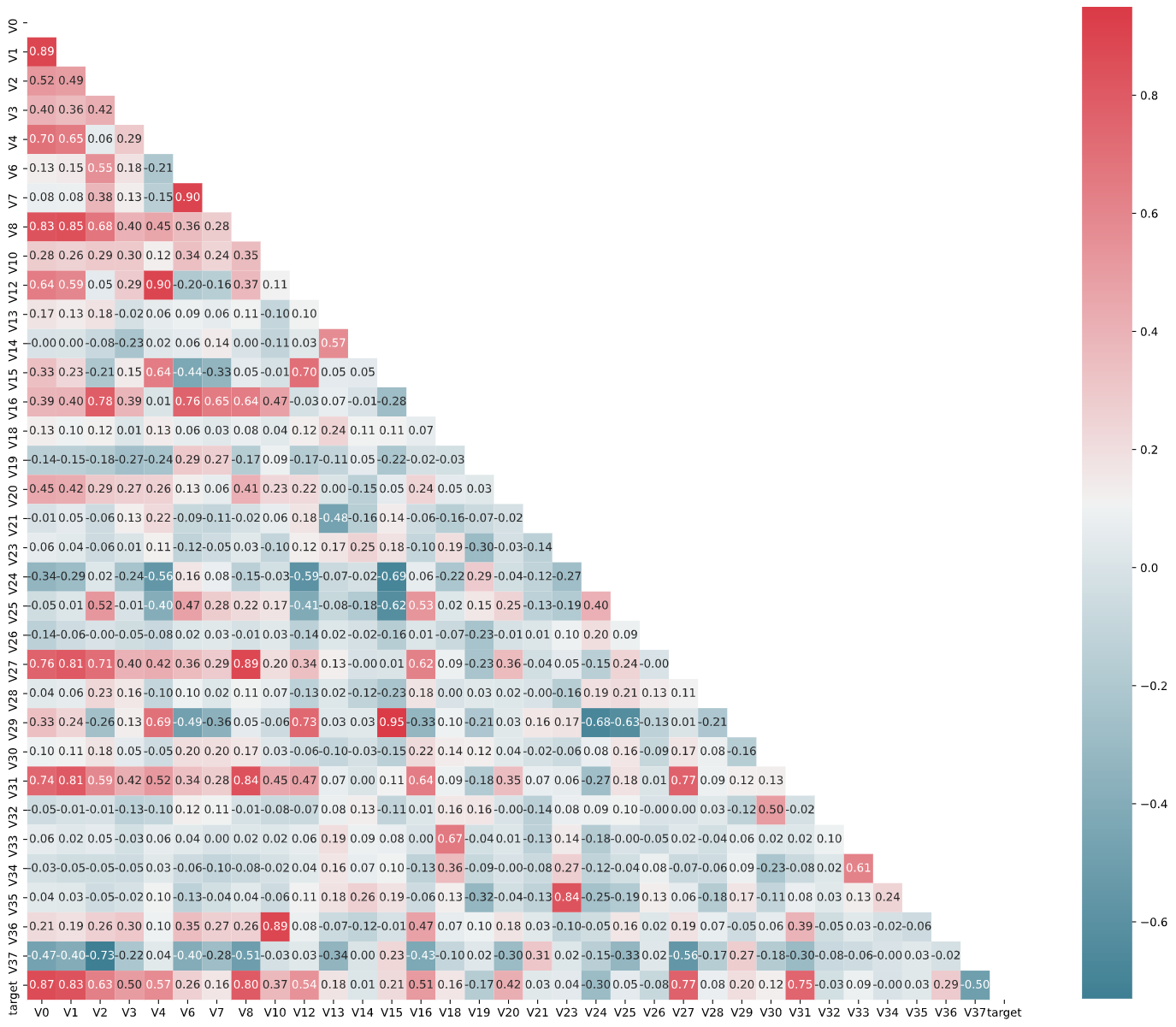
接下来进行查看特征之间的相关性(相关程度)。
1 2 3 4 5 6 7 8 9 data_train1 = data_all[data_all['oringin' ] == 'train' ].drop('oringin' , axis=1 ) plt.figure(figsize=(20 , 16 )) col = data_train1.columns.tolist() mcorr = data_train1[col].corr(method='spearman' ) mask = np.zores_like(mcorr, dtype=np.bool ) mask[np.triu_indices_from(mask)] = True cmap = sns.diverging_palette(220 , 10 , as_cmap=True ) g = sns.heatmap(mcorr, mask=mask, cmap=cmap, square=True , annot=True , fmt='0.2f' ) plt.show()
输出:
接下来继续进行降维操作。将和目标变量——蒸气值(target)相关性的绝对值小于阈值的特征进行删除。
1 2 3 4 5 threshold = 0.1 corr_matrix = data_train1.corr().abs () print('corr_matrix:' , corr_matrix) drop_col = corr_matrix[corr_matrix['target' ] < threshold].index data_all.drop(drop_col, axis=1 , inplace=True )
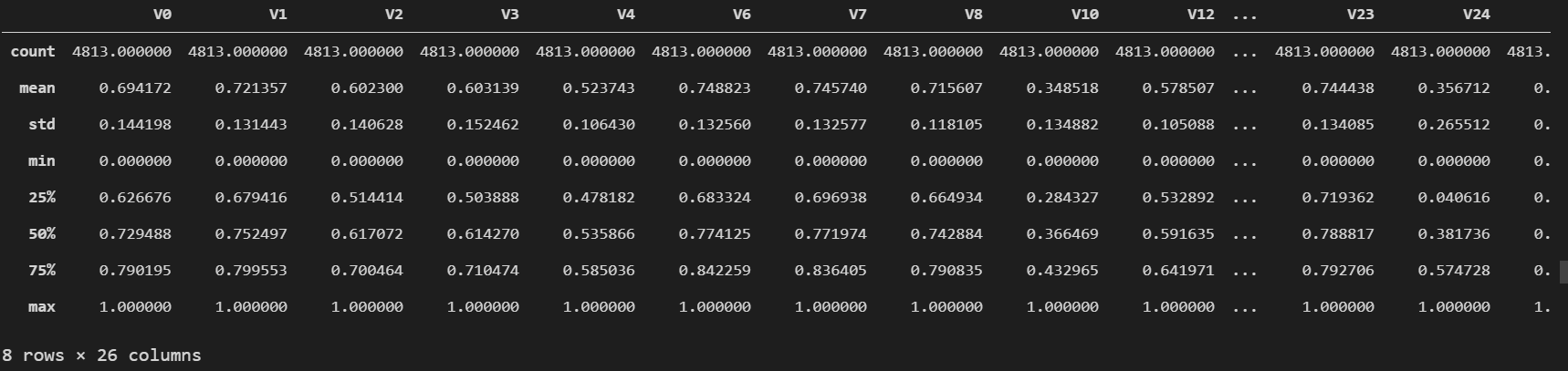
进行归一化 操作:
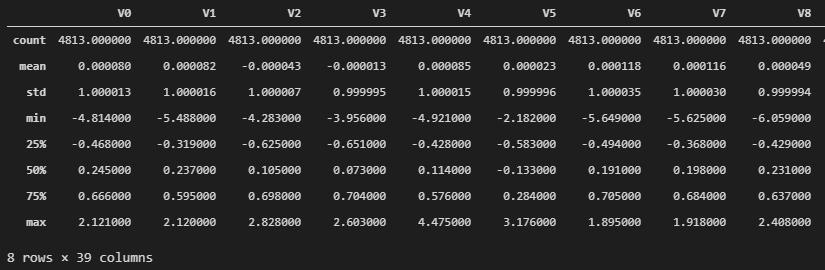
1 2 3 4 5 6 7 8 9 cols_numeric = list (data_all.columns) cols_numeric.remove('oringin' ) def scale_minmax (col ): return (col-col.min ()) / (col.max ()-col.min ()) scale_cols = [col for col in cols_numeric if col != 'target' ] data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax, axis=0 ) data_all[scale_cols].describe()
4.4 特征工程
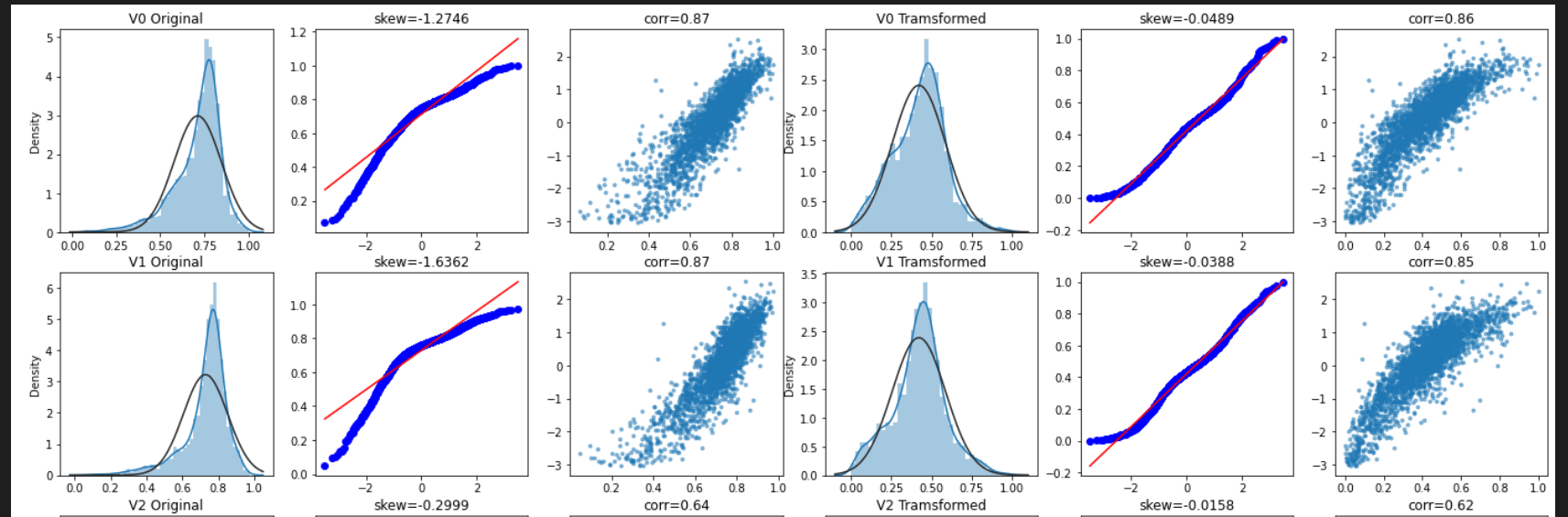
进行Box-Cox变换前,首先来看一下该变换对数据分布的影响。
关于Box-Cox变换可以参考:(传送门)[https://wenku.baidu.com/view/96140c8376a20029bd642de3.html]
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 fcols = 6 frows = len (cols_numeric) - 1 plt.figure(figsize=(4 *fcols, 4 *frows)) i = 0 for var in cols_numeric: if var != 'target' : dat = data_all[[var, 'target' ]].dropna() i += 1 plt.subplot(frows, fcols, i) sns.distplot(dat[var], fit=stats.norm) plt.title(var+' Original' ) plt.xlabel('' ) i+=1 plt.subplot(frows,fcols,i) _=stats.probplot(dat[var], plot=plt) plt.title('skew=' +'{:.4f}' .format (stats.skew(dat[var]))) plt.xlabel('' ) plt.ylabel('' ) i+=1 plt.subplot(frows,fcols,i) plt.plot(dat[var], dat['target' ],'.' ,alpha=0.5 ) plt.title('corr=' +'{:.2f}' .format (np.corrcoef(dat[var], dat['target' ])[0 ][1 ])) i+=1 plt.subplot(frows,fcols,i) trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1 ) trans_var = scale_minmax(trans_var) sns.distplot(trans_var , fit=stats.norm) plt.title(var+' Tramsformed' ) plt.xlabel('' ) i+=1 plt.subplot(frows,fcols,i) _=stats.probplot(trans_var, plot=plt) plt.title('skew=' +'{:.4f}' .format (stats.skew(trans_var))) plt.xlabel('' ) plt.ylabel('' ) i+=1 plt.subplot(frows,fcols,i) plt.plot(trans_var, dat['target' ],'.' ,alpha=0.5 ) plt.title('corr=' +'{:.2f}' .format (np.corrcoef(trans_var,dat['target' ])[0 ][1 ])) plt.show()
部分输出截图:
在进行Box-Cox变换之后,可以一定程度上减小不可观测的误差和预测变量的相关性。
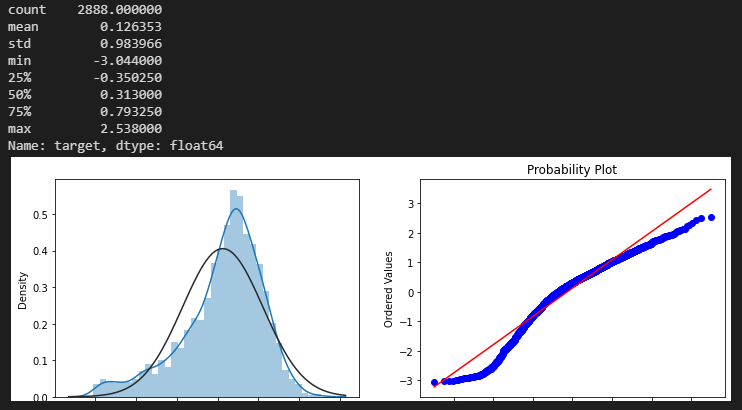
1 2 3 4 5 6 7 8 9 10 11 cols_transform = data_all.columns[0 :-2 ] for col in cols_transform: data_all.loc[:, col], _ = stats.boxcox(data_all.loc[:, col]+1 ) print(data_all.target.describe()) plt.figure(figsize=(12 , 4 )) plt.subplot(1 , 2 , 1 ) sns.distplot(data_all.target.dropna(), fit=stats.norm) plt.subplot(1 , 2 , 2 ) _ = stats.probplot(data_all.target.dropna(), plot=plt) plt.show()
输出:
可对target目标值使用对数变换,提升正态性。
1 2 3 4 5 6 7 8 sp = data_train.target data_train.target1 = np.power(1.5 , sp) plt.figure(figsize(12 , 4 )) plt.subplot(1 , 2 , 1 ) sns.distplot(data_train.target1.dropna(), fit=stats.norm) plt.subplot(1 , 2 , 2 ) _ = stats.probplot(data_train.target1.dropna(), plot=plt)
输出:
4.5 模型构建以及集成学习
1 2 3 4 5 6 7 8 9 10 11 12 def get_training_data (): df_train = data_all[data_all['oringin' ]=='train' ] df_train['label' ] = data_train.target1 y = df_train.target X = df_train.drop(['oringin' , 'target' , 'label' ], axis=1 ) X_train, X_valid, y_train, y_valid = train_test_split(X, y, test_size=0.3 , random_state=100 ) return X_train, X_valid, y_train, y_valid def get_test_data (): df_test = data_all[data_all['oringin' ] == 'test' ].reset_index(drop=True ) return df_test.drop(['oringin' , 'target' ], axis=1 )
————
RMSE:Root Mean Squard Error,均方根误差,是MSE(均方误差)开根号得到的值。
1 2 3 4 5 6 7 8 9 def rmse (y_true, y_pred ): diff = y_pred - y_true sum_sq = sum (diff**2 ) n = len (y_pred) return np.sqrt(sum_sq/n) def mse (y_true, y_pred ): return mean_squared_error(y_true, y_pred)
————
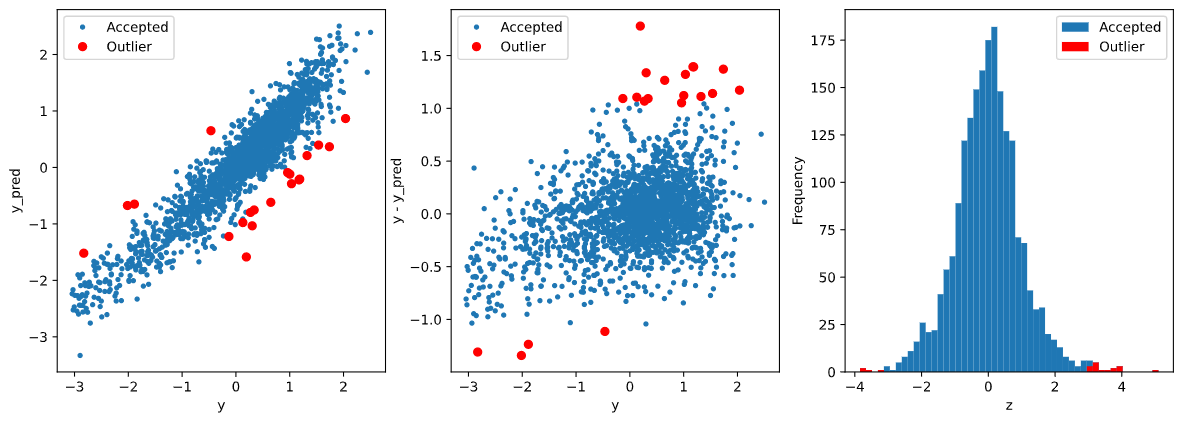
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 def find_outliers (model, X, y, sigma=3 ): model.fit(X, y) y_pred = pd.Series(model.predict(X), index=y.index) resid = y - y_pred mean_resid = resid.mean() std_resid = resid.std() z = (resid - mean_resid)/std_resid outliers = z[abs (z)>sigma].index print('R2=' , model.score(X, y)) print('rmse=' , rmse(y, y_pred)) print('mse=' , mse(y, y_pred)) print('---------------------------------------' ) print('mean of residual:' , mean_resid) print('std of residual:' . std_resid) print('---------------------------------------' ) print(len (outliers), 'outliers:' ) print(outliers.tolist()) plt.figure(figsize(15 , 5 )) ax_131 = plt.subplot(1 , 3 , 1 ) plt.plot(y, y_pred, '.' ) plt.plot(y.loc[outliers], y_pred.loc[outliers], 'ro' ) plt.legend(['Accepted' , 'Outliers' ]) plt.xlabel('y' ) plt.ylabel('y_pred' ) ax_132=plt.subplot(1 ,3 ,2 ) plt.plot(y,y-y_pred,'.' ) plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro' ) plt.legend(['Accepted' ,'Outlier' ]) plt.xlabel('y' ) plt.ylabel('y - y_pred' ) ax_133=plt.subplot(1 ,3 ,3 ) z.plot.hist(bins=50 ,ax=ax_133) z.loc[outliers].plot.hist(color='r' ,bins=50 ,ax=ax_133) plt.legend(['Accepted' ,'Outlier' ]) plt.xlabel('z' ) return outliers
1 2 3 4 5 6 7 8 X_train, X_valid, y_train, y_valid = get_training_data() test = get_test_data() outliers = find_outliers(Ridge(), X_train, y_train) X_outliers = X_train.loc[outliers] y_outliers = y_train.loc[outliers] X_t = X_train.drop(outliers) y_t = y_train.drop(outliers)
输出: 1 2 3 4 5 6 7 8 9 R2= 0.8767013861653619 rmse= 0.3489631754674053 mse= 0.12177529783229514 --------------------------------------- mean of residuals: -6.172422515926897e-16 std of residuals: 0.3490495418034555 --------------------------------------- 20 outliers: [2655, 2159, 1164, 2863, 1145, 2697, 2528, 2645, 691, 1874, 2647, 884, 2696, 2668, 1310, 1458, 2769, 2002, 2669, 1972]
1 2 3 4 def get_training_data_omitOutliers (): y = y_t.copy() X = X_t.copy() return X, y
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 def train_model (model, param_grid=[], X=[], y=[], splits=5 , repeats=5 ): if len (y) == 0 : X, y = get_training_data_omitOutliers() rkfold = RepeatedKFold(n_splits=splits, n_repeats=repeats) if len (param_grid) > 0 : gsearch = GridSearchCV(model, param_grid, cv=rkfold, scoring='neg_mean_squared_error' , verbose=1 , return_train_score=True ) gsearch.fit(X, y) model = gsearch.best_estimator_ best_idx gsearch.best_index_ grid_results = pd.DataFrame(gsearch.cv_results_) cv_mean = abs (grid_results.loc[best_idx, 'mean_test_score' ]) cv_std = grid_results.loc[best_idx, 'std_test_score' ] else : grid_results = [] cv_results = cross_val_score(model, X, y, scoring='neg_mean_squared_error' , cv=rkfold) cv_mean = abs (np.mean(cv_results)) cv_std = np.std(cv_results) cv_score = pd.Series({'mean' :cv_mean, 'std' :cv_std}) y_pred = model.predict(X) print('----------------------' ) print(model) print('----------------------' ) print('score=' ,model.score(X,y)) print('rmse=' ,rmse(y, y_pred)) print('mse=' ,mse(y, y_pred)) print('cross_val: mean=' ,cv_mean,', std=' ,cv_std) y_pred = pd.Series(y_pred, index=y.index) resid = y - y_pred mean_resid = resid.mean() std_resid = resid.std() z = (resid - mean_resid)/std_resid n_outliers = sum (abs (z)>3 ) outliers = z[abs (z)>3 ].index return model, cv_score, grid_results
1 2 3 4 5 opt_models = dict () score_models = pd.DataFrame(columns=['mean' , 'std' ]) splits = 5 repeats = 5
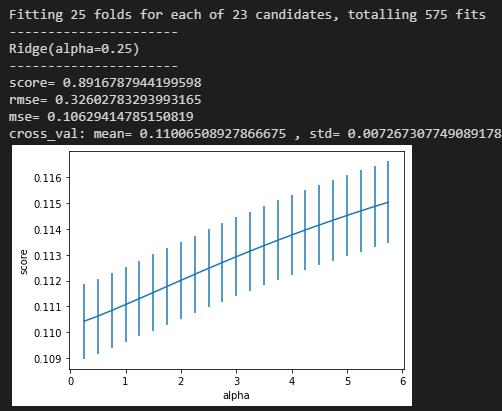
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 model = 'Ridge' opt_models[model] = Ridge() alph_range = np.arange(0.25 , 6 , 0.25 ) param_grid = {'alpha' :alph_range} opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid, splits=splits, repeats=repeats) cv_score.name = model score_models = score_models.append(cv_score) plt.figure() plt.errorbar(alph_range, abs (grid_results['mean_test_score' ]), abs (grid_results['std_test_score' ])/np.sqrt(splits*repeats)) plt.xlabel('alpha' ) plt.ylabel('score' ) plt.show()
输出:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 def model_predict (test_data, test_y=[] ): i = 0 y_predict_total = np.zeros((test_data.shape[0 ],)) for model in opt_models.keys(): if model != 'LinearSVR' and model != 'KNeighbors' : y_predict = opt_models[model].predict(test_data) y_predict_total += y_predict if len (test_y) > 0 : print('{}_mse:' .format (model), mean_squared_error(y_predict, test_y)) y_predict_mean = np.round (y_predict_total/i, 6 ) if len (test_y) > 0 : print("mean_mse:" ,mean_squared_error(y_predict_mean,test_y)) else : y_predict_mean=pd.Series(y_predict_mean) return y_predict_mean
————
1 2 y_ = model_predict(test) y_.to_csv('predict.txt' , header=None , index=False )